Below is a summary of the research findings of PRISMS funded post-doctoral fellow, Takao Tsukahara D.D.S Ph.D. Takao was funded by PRISMS from January 2018 to March 2020. He will be continuing his work on RAI1 at the University of Michigan. We are excited to share his current findings with our community, and look forward to his future research!
Takao Tsukahara D.D.S Ph.D
Drs. Iwase and Sutton laboratories
University of Michigan
Summary
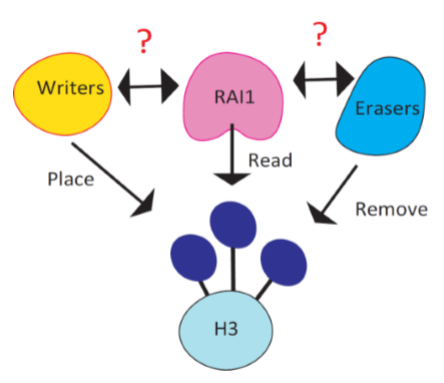
It is reported that Rai1, the causal gene of Smith Magenis syndrome (SMS), reads the methylation code placed at histone 3 lysine 4 (H3K4me). Consistent to Rai1’s role in gene regulation, H3K4me is known as a mark of transcriptionally active area in the genome. H3K4me is placed by six writers (KMT2A-G) and removed by six erasers (KDM1A-B, KDM5A-D) and loss of function in these enzymes are mostly associated with neurodevelopmental disorders. Therefore, it is likely that some or one of these enzymes share common pathways with Rai1.
The goal of this project is to screen the roles of H3K4me enzymes in synaptic scaling, an enduring form of synaptic plasticity, and further look at gene expression programs regulated by these enzymes to pinpoint targets that interacts with Rai1’s functional pathways.
Recent findings from our group unraveled that Rai1 has at least two functions in maintaining stabilized neural network. Firstly, Rai1 optimize synaptic strength in a resting state. Secondly, Rai1 mediates the gene expression program driven by reduced network activity, thereby strengthening synapse to stabilize network activity.
Using RNA interference, genetic deletion and chemical inhibitors, we characterized all six writers. Interestingly, KMT2C/D and KMT2A/B showed similar and opposing function to Rai1, respectively. These results suggest that KMT2C/D might share some common pathways with Rai1 and KMT2A/B might functionally counter with Rai1. We are continuously screening eraser enzymes and plan to reveal the gene expression programs regulated by these enzymes to further understand the relationship with Rai1
Method
To systematically test the roles of individual H3K4 enzymes in synaptic scaling, without the impact of gene manipulation in synapse development, we first employed an RNAi approach. We co-transfected shRNA-expression plasmids (all validated by RT-PCR) with a GFP-expression plasmid in primary hippocampal neurons (12-14 days in vitro, DIV), which targets roughly 1% of neurons allowing for an analysis of cell-autonomous roles of KMT2 enzymes. We then induced synaptic upscaling or downscaling through the application of the Na+ channel blocker tetrodotoxin (TTX; 1 µM, 24 hrs) or the GABAA receptor antagonist bicuculline (Bic; 20 µM, 24 hrs), respectively.
Results
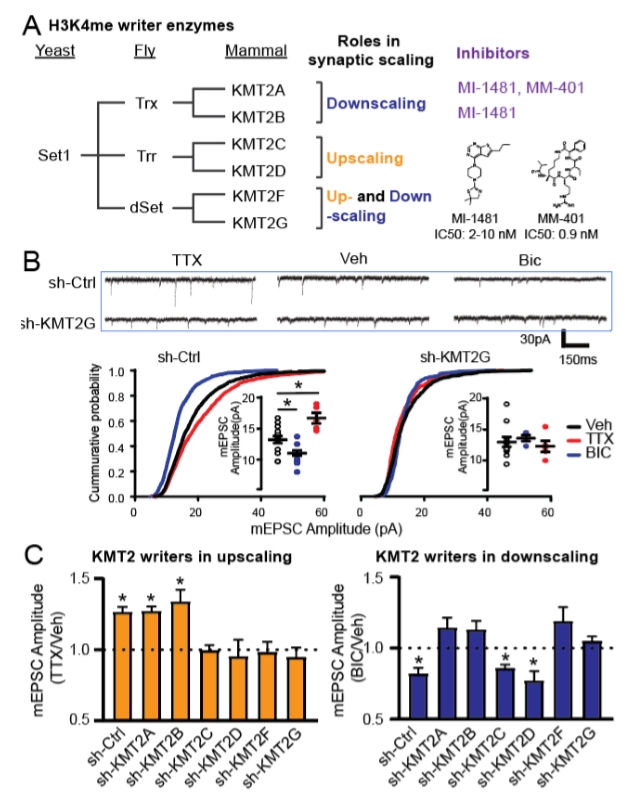
Neurons transfected with control scrambled shRNA showed robust increases and decreases, respectively, in the amplitude of miniature excitatory post-synaptic currents (mEPSCs) in response to chronic TTX and Bic. Remarkably, individual knockdown of all of the six KMT2 enzymes led to distinct outcomes in synaptic scaling. Knockdown of KMT2A and KMT2B blocked synaptic downscaling induced by Bic, while depletion of KMT2C and KMT2D interfered with upscaling triggered by TTX (Fig. 1). Knockdown of KMT2F and KMT2G blocked both up- and downscaling.
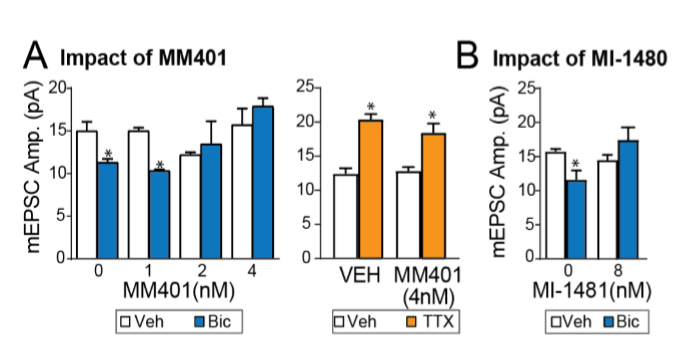
MM401, the KMT2A specific inhibitor, as well as the KMT2A/B inhibitor MI-1481 also prevented synaptic downscaling, without affecting upscaling (Fig. 2)
Future Directions
We are further delineating the role of H3K4me enzymes and if we could find a specific enzyme that strongly interacts or counters with Rai1’s functional pathway, that enzyme will become an attractive target to intervene with in SMS.